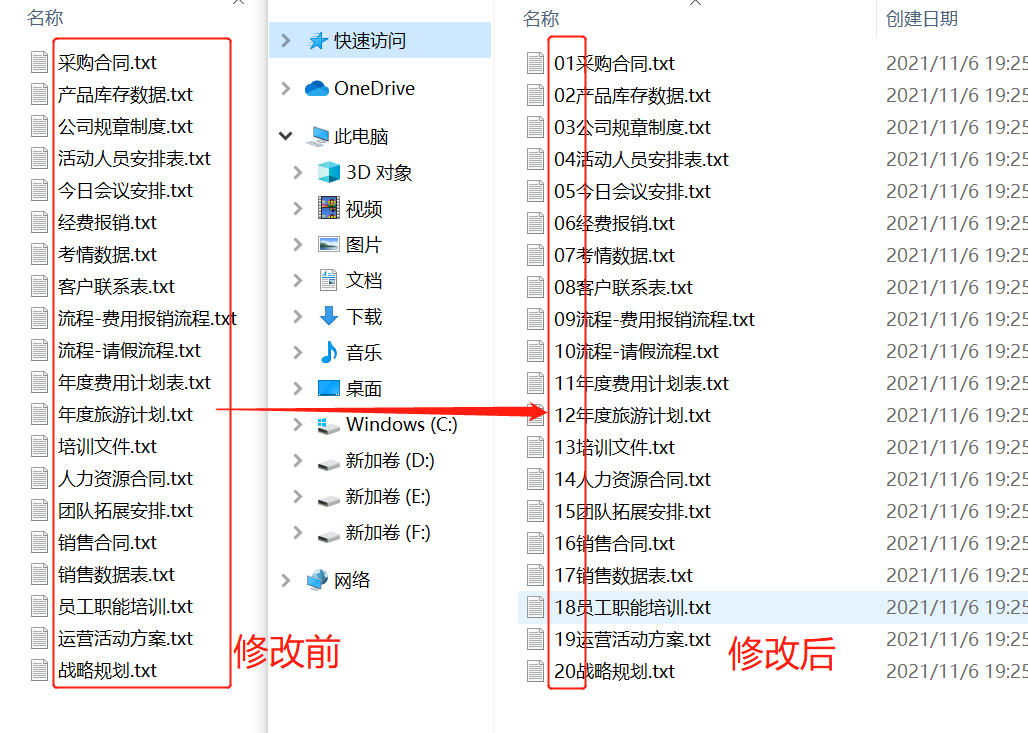
Python脚本:PDB文件中原子和残基重新编号
Command:
python renumber_pdb.py -i protein.pdb -a -r > output.pdb
renumber_pdb.py
# Python 3 script to atoms and residues in a PDB file.
#
# run
# ./renumber.py -h
# for help
#class Pdb(object):""" Object that allows operations with protein files in PDB format. """def __init__(self, file_cont = [], pdb_code = ""):self.cont = []self.atom = []self.hetatm = []self.fileloc = ""if isinstance(file_cont, list):self.cont = file_cont[:]elif isinstance(file_cont, str):try:with open(file_cont, 'r') as pdb_file:self.cont = [row.strip() for row in pdb_file.read().split('\n') if row.strip()]except FileNotFoundError as err:print(err)if self.cont:self.atom = [row for row in self.cont if row.startswith('ATOM')]self.hetatm = [row for row in self.cont if row.startswith('HETATM')]self.conect = [row for row in self.cont if row.startswith('CONECT')]def renumber_atoms(self, start=1):""" Renumbers atoms in a PDB file. """out = list()count = startfor row in self.cont:if len(row) > 5:if row.startswith(('ATOM', 'HETATM', 'TER', 'ANISOU')):num = str(count)while len(num) < 5:num = ' ' + numrow = '%s%s%s' %(row[:6], num, row[11:])count += 1out.append(row)return outdef renumber_residues(self, start=1, reset=False):""" Renumbers residues in a PDB file. """out = list()count = start - 1cur_res = ''for row in self.cont:if len(row) > 25:if row.startswith(('ATOM', 'HETATM', 'TER', 'ANISOU')):next_res = row[22:27].strip() # account for letters in res., e.g., '1A'if next_res != cur_res:count += 1cur_res = next_resnum = str(count)while len(num) < 3:num = ' ' + numnew_row = '%s%s' %(row[:23], num)while len(new_row) < 29:new_row += ' 'xcoord = row[30:38].strip()while len(xcoord) < 9:xcoord = ' ' + xcoordrow = '%s%s%s' %(new_row, xcoord, row[38:])if row.startswith('TER') and reset:count = start - 1out.append(row)return outif __name__ == '__main__':import argparseparser = argparse.ArgumentParser(description='Renumber residues in a pdb file',formatter_class=argparse.RawTextHelpFormatter)parser.add_argument('-i', '--input', help='Input PDB file')parser.add_argument('-s', '--start', help='Number of the first residue in the renumbered file (default = 1)')parser.add_argument('-a', '--atoms' ,action='store_true', help='Renumbers atoms')parser.add_argument('-r', '--residues', action='store_true', help='Renumbers residues')parser.add_argument('-c', '--chainreset', action='store_true', help='Resets the residue renumbering after encountering a new chain.')parser.add_argument('-v', '--version', action='version', version='v. 1.0')args = parser.parse_args()if not args.input:print('{0}\nPlease provide an input file.\n{0}'.format(50* '-'))parser.print_help()quit()if not args.start:start = 1else:start = int(args.start)if not args.atoms and not args.residues:print('{0}\nPlease provide at least the --atoms or --residues flag.\n{0}'.format(50* '-'))parser.print_help()quit()pdb1 = Pdb(args.input)if args.atoms:pdb1.cont = pdb1.renumber_atoms(start=start)if args.residues:pdb1.cont = pdb1.renumber_residues(start=start, reset=args.chainreset)for line in pdb1.cont:print(line)
效果:
参考:
https://github.com/AspirinCode/protein-science/tree/master/scripts-and-tools/renumber_pdb